Metabolic Network Analysis of an Anaerobic Microbial Community: Potential for Syntrophic Methane and Hydrogen Production
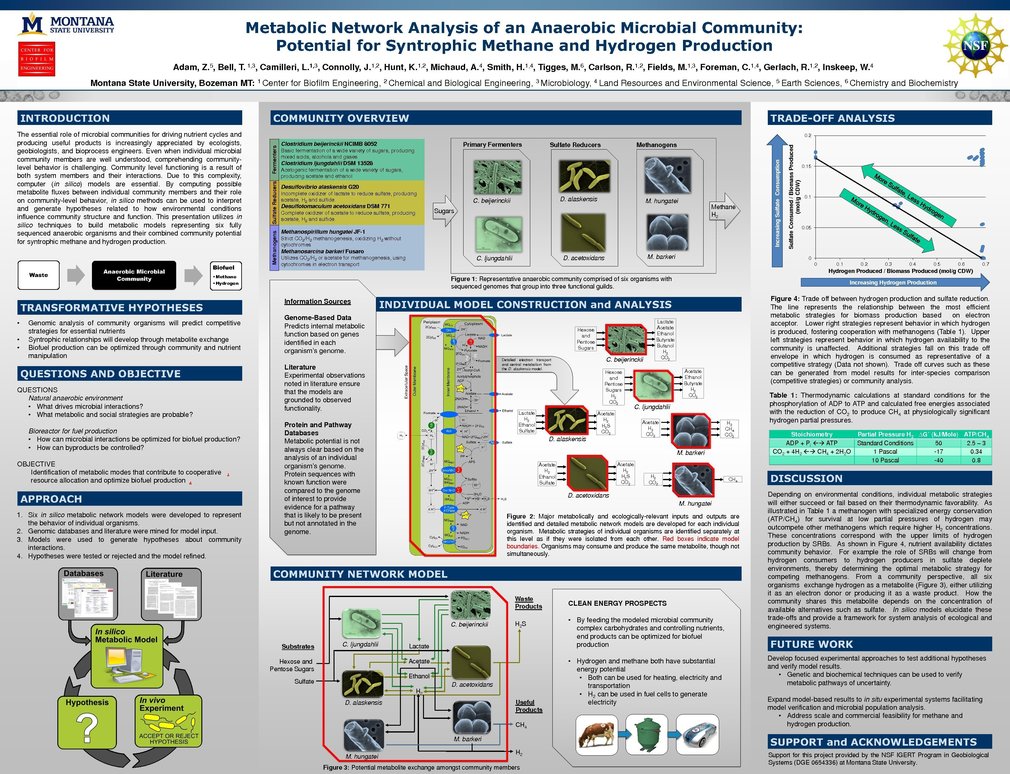
Microbial community interactions represent a research area of growing interest to many scientific and engineering disciplines, including ecologists, geobiologists and bioprocess engineers. Community-level behavior is a complex result of both system members and their interactions, which can complicate the development of testable hypotheses for ex vivo microbial systems even when individual microbial community components are well understood. In the absence of suitable natural analogs, computer (in silico) models of interactions between distinct microbial metabolic groups can assist the characterization of complex microbial ecosystem behavior. Due to increases in sequencing speed and decreases in sequencing costs, relevant genomic data for many microbial systems are readily available. This genomic information can be translated into collections of biochemical reactions that capture the organism’s metabolic potential, permitting a “systems based” analysis of relationships between environmental parameters, genome/metagenome content and community functioning. By assessing metabolite flux between individual community members and their effects on community-level behavior, in silico methods can be used to generate hypotheses related to environmental conditions and community structures that achieve useful outcomes. These outcomes include understanding and optimizing both natural and industrial processes of pressing societal importance, such as increased biofuel production or more efficient waste remediation. This presentation discusses in silico techniques used to build models representing six fully sequenced anaerobic organisms, and their combined community potential for predicting syntrophic methane and hydrogen production.
Metabolic Network Analysis of an Anaerobic Microbial Community: Potential for Syntrophic Methane and Hydrogen Production
Microbial community interactions represent a research area of growing interest to many scientific and engineering disciplines, including ecologists, geobiologists and bioprocess engineers. Community-level behavior is a complex result of both system members and their interactions, which can complicate the development of testable hypotheses for ex vivo microbial systems even when individual microbial community components are well understood. In the absence of suitable natural analogs, computer (in silico) models of interactions between distinct microbial metabolic groups can assist the characterization of complex microbial ecosystem behavior. Due to increases in sequencing speed and decreases in sequencing costs, relevant genomic data for many microbial systems are readily available. This genomic information can be translated into collections of biochemical reactions that capture the organism’s metabolic potential, permitting a “systems based” analysis of relationships between environmental parameters, genome/metagenome content and community functioning. By assessing metabolite flux between individual community members and their effects on community-level behavior, in silico methods can be used to generate hypotheses related to environmental conditions and community structures that achieve useful outcomes. These outcomes include understanding and optimizing both natural and industrial processes of pressing societal importance, such as increased biofuel production or more efficient waste remediation. This presentation discusses in silico techniques used to build models representing six fully sequenced anaerobic organisms, and their combined community potential for predicting syntrophic methane and hydrogen production.
10007 Views
-
Further posting is closed as the event has ended.

Judges and Presenters may log in to read queries and replies.